|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Here you will find informations on the materials, methods, and quality controls applied to generate the Redox Atlas of the Mouse. optimized for Mozilla Firefox at >= 1200x800 resolution |
Antibody sources The majority of the antibodies used in this study (Trx1, TrxR1, Trx2, Grx1, Grx2, Grx3, Grx5, Prx1, Prx3, and Prx5) are polyclonal antibodies raised in rabbits against recombinantly expressed proteins. The antibodies detecting γ-GCS (i.e. the catalytic subunit GCLC), Prx2, Prx4, Prx6, and TrxR2, were obtained from commercial vendors as listed below. In addition, we have tested alternative antibodies for two of the antigens, Trx1 (ATRX-06, IMCO Corp. Ltd. AB, Stockholm Sweden) and TrxR2 (ab58445, Abcam plc, Cambridge, UK). These resulted, with only minor differences in intensities, in essentially identical stainings when compared to the antibodies used throughout the rest of our study.
Antibody validation Antibodies and sera were evaluated by Western blotting and immunohistochemistry comparing antibody-stained samples with samples stained with antigen-saturated antibodies (see figures 1 and 2A). Only antibody preparations that lost immunoreactivity by the blocking procedure and displayed specific bands of the expected size in Western blotting were used in this study. 50 % of the initially produces sera fulfilled this requirement. Two sera (raised against mouse Grx3 and rat TrxR1) were additionally affinity purified using immobilized antigen until the requirements described above were met. The specificity of the antibodies obtained from commercial vendors was confirmed by Western blotting (figure 2B). 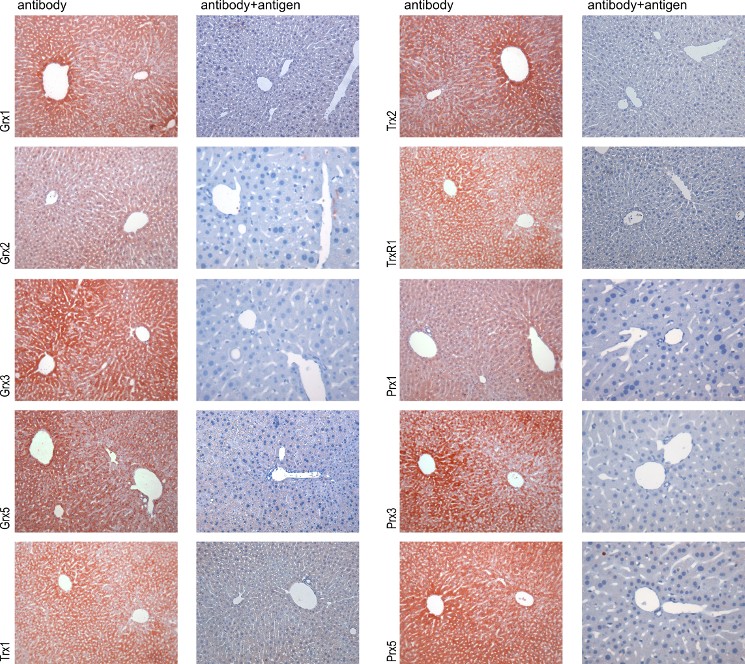
Figure 1 - Validation of the antibodies by immunohistochemistry A comparison of antibody-stained liver samples (left side) with samples stained with antibodies blocked with 10-200 µg/ml of the specific antigen (right side) is shown for each antigen. The disappearance of staining by blocking with the specific antigen was used as indication for the specificity of the antibodies/sera for the antigen. 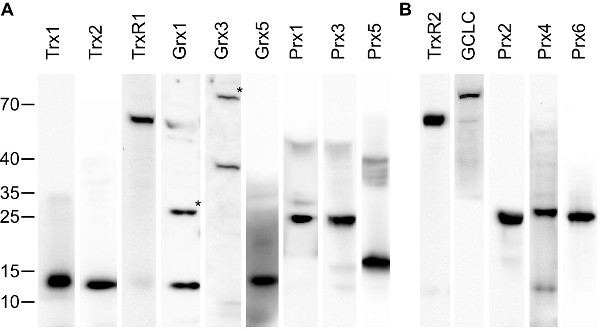
Figure 2 – Validation of the antibodies by Western blotting. 30 µg of mouse liver extracts were separated by SDS Page, blotted on PVDF membranes, and incubated with the specific antibodies/sera. Subsequently, antibodies were detected with HRP-coupled anti-rabbit antibodies and enhanced chemiluminescence staining. (A) Antibodies produced in this study, (B) antibodies from commercial vendors. All antibodies resulted in one major band with no or only minor additional bands visible. (*) Grx1 and Grx3 displayed two bands, from which the higher (of approx. two-times the molecular weight of the lower one) disappeared upon extensive reduction of the extracts with DTT (not shown). Hence, the two bands correspond to the monomeric and (disulfide-bridged) dimeric proteins. GCLC is the catalytic subunit of γ-GCS. The specificity of the anti-Grx2 antibodies in Western blotting experiments have been demonstrated before [Hudemann et al. 2009]. Animals and tissue preparation Mice of the strain 129 were sacrificed by cervical dislocation. Organs of interest were quickly removed, segmented (when necessary), and placed in 4 % para-formaldehyde solution for fixation at 4 °C for 24 h. For brain dissections, mice (age 6 month - 1 year) were deeply anesthetized with sodium-pentobarbital (Narcoren, Merial, Hallbergmoos, Germany) by a dose of 100 mg/kg intraperitoneally and transcardially perfused with 0.9 % NaCl followed by the fixation solution (4 % para-formaldehyde in 0.1 M sodium phosphate buffer). Brain pieces were postfixed in the same fixative at 4 °C for 24 h. Following dehydration, the samples were processed for paraffin embedding. Sections (5-10 µm thick) were cut and placed on poly-L-lysine-coated or silanized slides. Before staining, sections were deparaffinized and incubated in 3 % hydrogen peroxide for 10 min to quench endogenous peroxidase. Immunohistochemistry Nons-pecific antibody binding sites were blocked with 10 % normal goat serum (PAA) in PBS. Next, sections were incubated overnight with the primary antibodies diluted in blocking solution at 4 °C. Negative controls were incubated with 10 % goat serum in PBS. The following day, sections were washed 3 times with PBS and subsequently incubated with a biotinylated goat anti-rabbit antibody diluted 1:500 (DAKO, Glostrup, Denmark) for 60 min at room temperature. The HRP streptavidin detection system (KPL, Gaithersburg, USA) was used for antigen staining according to the manufacturer's recommendations. Sections were incubated with the substrate aminoethyl carbazole (AEC, Invitrogen, Karlsruhe, Germany) for 5 min at room temperature, counter-stained with Mayer's hematoxylin and mounted with Mowiol. Sections were examined with a Leica Diaplan microscope equipped with a MicroPublisher camera (QImaging, Surrey, BC, Canada). The white balance was adjusted using the GNU image manipulation program (www.gimp.org), the panels were arranged using inkscape (www.inksacape.org). No additional image editing was performed. Quality assurance In addition to the validation of the antibodies described above, we have excluded the possibility of background staining (derived from the secondary antibodies) for all specimen by including control stainings in every staining series in which the primary antibodies were omitted. Each pictures represents up to 5 independent stainings with essential identical features. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||