|
||||
The links provided to the right will lead you to additional resources regarding the individual redox proteins. optimized for Mozilla Firefox at >= 1200x800 resolution |
The redox proteins studied in this work In this study, we have analyzed the distribution of some prominent members of the thioredoxin family of proteins:
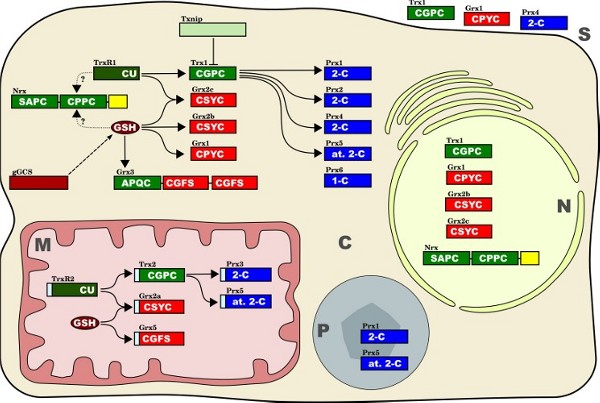
Figure 3 - Isoforms, subcellular localization and confirmed interactions between the thiol redox proteinsof the thioredoxin family investigated in this study. The active site sequences and the classes of redoxins, respectively, are indicated in white. The one-letter code for amino acids was used to identify active site sequences, for instance CU = cysteine-selenocysteine. Thioredoxin domains (and related proteins) are shown in green, glutaredoxin domains (and related proteins/compounds) in red and brown, peroxiredoxins in blue. Peroxiredoxin classes: 1-C = one Cys Prx, 2-C = two Cys Prx, at. 2-C = atypical two Cys Prx. Straight arrows indicate potential electron transfer reactions between the different redox proteins. Dotted arrows indicate putative electron transfer reactions. Txnip (thioredoxin interacting protein) is regarded as natural inhibitor of thioredoxin function. The dashed arrow indicates the involvement of gGCS in the synthesis of glutathione (GSH). Cellular compartments: C, cytosol; M, mitochondria; N, nucleus; P, peroxisomes; S, secreted. |
 |
||